






 Introduction
Introduction
 How to use MultAlin
How to use MultAlin
 Input formats
Input formats
 Output format
Output format
 Other parameters
Other parameters
 Presentation options
Presentation options
 More information
(complete MultAlin documentation)
More information
(complete MultAlin documentation)
 This server history
This server history

 Introduction
Introduction
Welcome to Multalin!
This software will allow you to align simultaneously several biological sequences.
What is a Multiple sequence alignment? It is the arrangement of several protein or nucleic acid sequences with postulated gaps so that similar residues are juxtaposed. A positive score is attached to identities, conservative or non-conservative substitutions (the score amplitude measuring the similarity) and a penalty to gaps; an ideal program would maximise the total score, taking account of all possible alignments and allowing for any length gap at any position.
Unfortunately the computing requirements, both of time and memory, grow as the nth power, where n is the sequence number, so this ideal alignment can be found only for two sequences or three short sequences. In the general case, to be practicable programs must restrict the conditions of the optimisation. Nevertheless it is undeniably useful to have an automatic system available for multiple sequence alignment to provide a starting point for a more human analysis.
Multalin creates a multiple sequence alignment from a group of related sequences using progressive pairwise alignments. The method used is described in "Multiple sequence alignment with hierarchical clustering", F.Corpet, 1988, Nucl. Acids Res. 16 10881-10890.



Warning : No computer skills are required to use MultAlin, only basic www knowledge !
On the MultAlin home page you will see a large rectangle. This is where you are going to paste (as in cut and paste) your sequences (try a sample set of sequences the first time). Instead of pasting your sequences, you can give the name of your sequences file, or select it with the Browse button.
The next step is to set the parameters. These are only of basic www difficulty but you will be able to find help by clicking on the associated question mark. Simply use the pop up menus or type in text or numbers where required. When you are ready click on the "submit data" button (you can use either the buttons at top or at bottom of the page .
Now you will have to wait for our server to calculate.(this can take up to a few hours for very large sequences).
The result will be sent back to your internet browser in the form of a GIF image (default), a plain text or a coloured html page. You will be able to change the colours, font size, line size etc. and even the consensus levels (see Presentation options for details).
The procedure is the same as for the MultAlin set-up, just use the pop up menus and type in text or numbers where required. When ready click on the "Apply Changes" button. The new image will appear shortly after. (only the image is changed, no realignment is done)
On your result page, you can add a sequence to the alignment. This sequence will be aligned with your already aligned sequences and you'll get a new result page, with the new sequence placed beside its more similar sequence. For this step, MultAlin performs an optimal alignment of the new sequence and the block of the already aligned sequences: the result can be different if you directly ask for an alignment of all the sequences in the first form.
Paste your new sequence in the rectangle aera in Fasta/Multalin format (i.e. one line with a beginning '>' for the sequence name, and other lines with rhe sequence itself). Click on the "Apply Changes" button when ready.



 MultAlin-Fasta
MultAlin-Fasta
The MultAlin format is similar to Fasta. Sequences can be interrupted by spaces
or digits not taken into account (see samples in MultAlin and pure Fasta formats)
> SeqName the sequence name is the
> first word of the first comment line
> max: 8 letters
> comment lines begin with >
AAAACCGTTAAA...
> SeqNam2 the 2nd sequence beginning
> shows the end of the first one
AAACCTGGAC...



 GenBank
GenBank
LOCUS SeqName
any lines
ORIGIN anything
1 aggtcccttt tgtgttgttt
The sequence name is the first word after the LOCUS key-word. The sequence begins on the line following the ORIGIN key-word. The next sequence information begins with the LOCUS key-word. See sample.



 EMBL-SwissProt
EMBL-SwissProt
ID SeqName
any lines
SQ anything
aauccagug gagaucaaag
any sequence lines
//
The sequence name is the first word after the ID key-word. The sequence begins on the line following the SQ key-word. The next sequence information begins on the line following // See sample.



 Output format
Output format
 The sequence alignment will be displayed as:
The sequence alignment will be displayed as:In any case you can adjust the consensus levels.
 Available
files
Available
filesJust underneath you will be able to see the input sequence file, the cluster file, the alignment in fasta or msf format plain text, the alignment in msf format with colour indications as a coded text, an html text or a gif image.
Any of these files can be saved to your local disk, simply using your WWW browser. The plain texts can be viewed, edited or printed with any text editor, the Html page and the GIF image, with your browser or a text processor that allows these formats.
To translate the colour indications of the coded text to true colours, you can use Microsoft Word and the MultAlin macro (FTP multalin.dot and save to disk even if you see odd characters on your browser) as follow:
Open your .doc file with Microsoft Word (File/Open) Change the templates (File/Models... or Tools/Models..., Link..., search the disk to select multalin.dot, Open) Run MultAlin Macro (Tools/Macro..., select MultAlin, Run) You can also add MultAlin macro to your current model (Normal.dot): Tools/Macro..., Organizer, Close File then Open File (on the same button), search the disk to select multalin.dot, Open, select MultAlin, Copy >> into Normal.dot, Close



 Other parameters
Other parameters
 Symbol comparison table
Symbol comparison table



 Gap penalties
Gap penalties



 Gap penalty at extremities
Gap penalty at extremities



 One iteration only
One iteration only

 Presentation options
Presentation options
 Text options
Text options
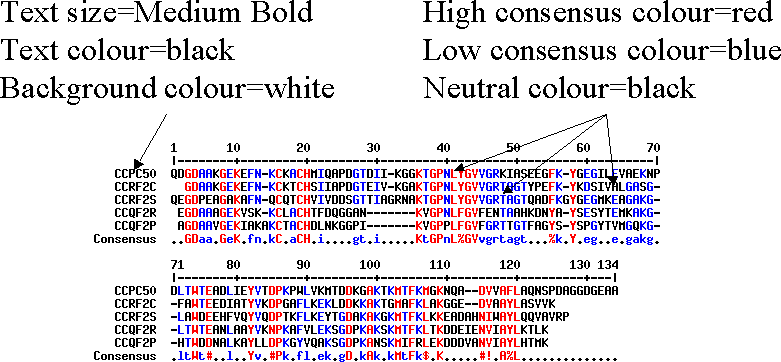
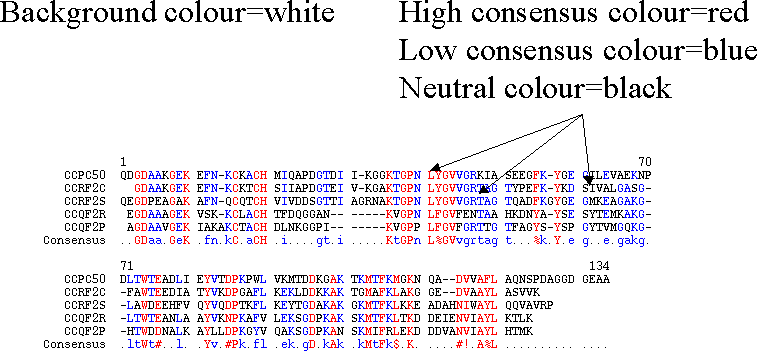
 Consensus options
Consensus options
 Other presentation options
Other presentation options
CCQF2P aGDAAvGEK iakaKCtACH dlnkggpi-- -----KvGPp LFGVfGRTtG TfagYs-Ysp GytvmGqKG-
Consensus ..GDaa.GeK .fn.kC.aCH .i....gt.i .....KtGPn L%GVvgrtag t...%k.Y.e g..e.gakg.
CCPC50 QDGDAAKGEK EFN-KCKACH MIQAPDGTDI I-KGGKTGPN LYGVVGRKIA SEEGFK-YGE GILEVAEKNP
CCRF2C ........ ...-...T.. S.I.....E. V-..A..... .......TAG TYPE..-.KD S.VALGASG-






 Florence Corpet
MultAlin's author. (Comments and suggestions very welcome)
Florence Corpet
MultAlin's author. (Comments and suggestions very welcome)